Research in the Xue Lab
Transcriptional programs as therapeutic targets in eradicating leukemic stem cells:
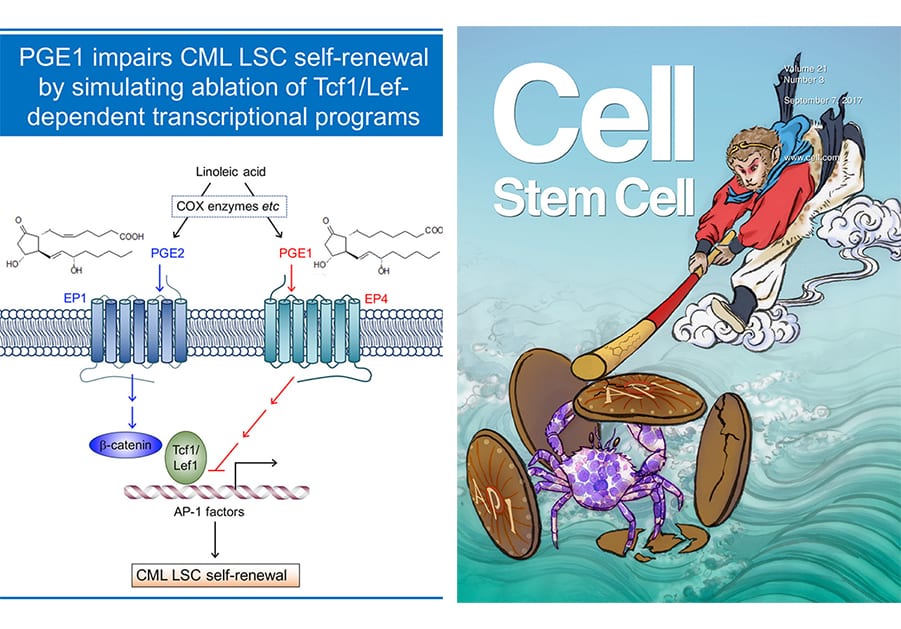
Hematopoietic and leukemic stem cells (HSCs and LSCs, respectively) are endowed with self-renewal capacity. HSCs sustain lifetime production of multiple blood lineages, and LSCs are responsible for initiation, maintenance, and propagation of leukemia, including chronic and acute myeloid leukemia (CML and AML). A cure for leukemia depends on the ability to eradicate LSCs after effective debulking of leukemic blasts with conventional chemotherapy. Our laboratory has been investigating key transcriptional programs that are differentially utilized in HSCs and CML LSCs (Yu et al, Blood, 2011 and Yu et al, Cell Stem Cell, 2012. Our recent efforts revealed that Tcf1 and Lef1 transcription factors are specifically required for self-renewal of CML LSCs but not normal HSCs , and identified two small molecules, prostaglandin E1 (PGE1, aka alprostadil) and its analogue misoprostol, that showed ability to simulate the impact of Tcf1/Lef1 gene ablation on CML LSCs (Li et al, Cell Stem Cell, 2017). This work appears as cover story on Cell Stem Cell, demonstrates the powerful potential and therapeutic values of molecular dissection of key regulatory factors/pathways in HSCs and LSCs. The Xue lab is currently applying this successful strategy to target LSCs of AMLs, which are genetically heterogeneous and characterized by a multitude of chromosomal abnormalities and somatic mutations.
To make a functional T cell and avoid malignant transformation:
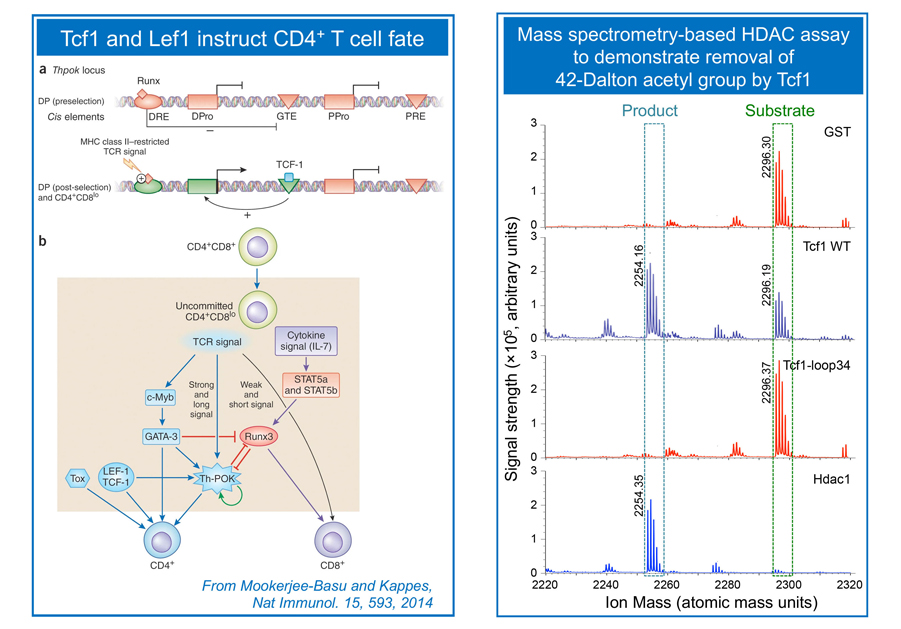
T cells develop in the thymus by following clearly defined stage-wise maturation. Early thymic progenitors (ETPs) are committed to the T cell lineage at the CD4–CD8– double negative (DN) stage. At this stage, TCRb locus is rearranged, which is associated with double-strand DNA breaks. We demonstrate that the transcription factor Tcf1 functions as a tumor suppressor in addition to contributing to T cell lineage specification (Yu et al. Immunity, 2012). CD4+CD8+ double positive (DP) thymocytes are bipotent, giving rise to CD4+ and CD8+ single positive (SP) cells in the helper and cytotoxic lineages, respectively. The lineage choice decision in DP thymocytes is instructed by a network of transcriptional regulators, and we showed that Tcf1 and its homologous protein, Lef1, act upstream to ThPOK to instruct DP cells to a CD4+ T cell fate (Steinke, Yu et al. Nat. Immunol. 2014). Following lineage choice decisions, the SP cells undergo intra-thymic maturation to cement their distinct T cell identity. We discovered that Tcf1 and Lef1 have intrinsic histone deacetylase (HDAC) activity, which is required for suppressing CD4+ lineage-associated genes to establish CD8+ T cell identity (Xing, Li et al, Nat. Immunol. 2016). This finding demonstrates that sequence-specific transcription factors have the capacity to directly modify histones. The Xue lab continues this line of investigation to dissect cooperation between transcriptional and epigenetic regulators in T cell lineage specification and T cell identity, while suppressing malignancy in developing T cells.
Mapping pathways that promote T follicular helper cell differentiation:
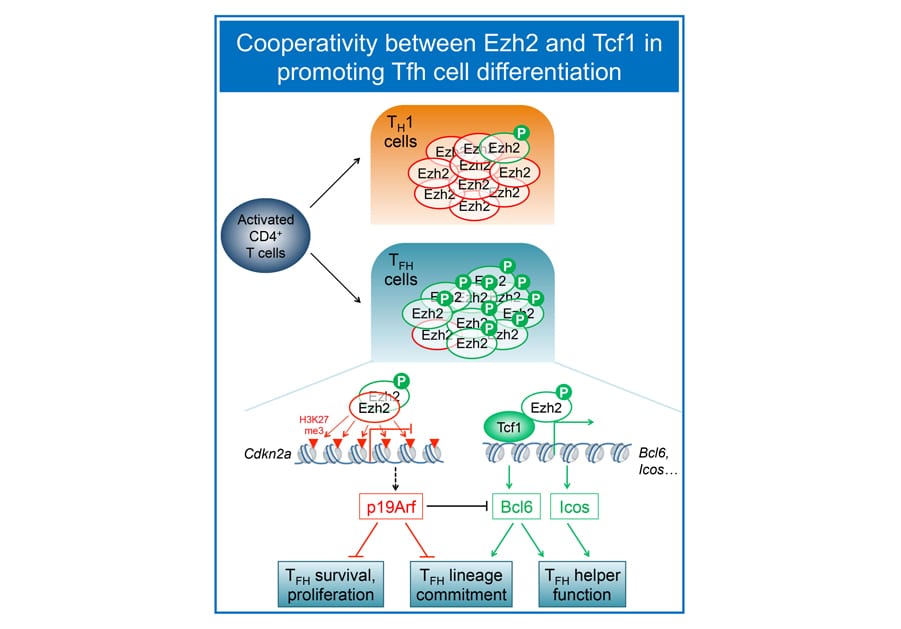
Mature CD4+ T cells provide versatile helper functions, and we are particularly interested in T follicular helper (Tfh) cells, which are specialized in help B cells to generate class-switched, high-affinity antibodies. Our collaborative work with the Crotty lab (LJI) demonstrated that Tcf1 and Lef1 act upstream of the Tfh lineage-defining transcription factor Bcl6, as well as ICOS and IL-6 receptor to promote Tfh cell differentiation (Choi, Gullicksrud et al, Nat. Immunol. 2015). Follow-up studies further reveal that 1) Tcf1 short isoforms, without the capacity to interact with b-catenin, are adequate to support Bcl6 transcriptional activation and promote Tfh differentiation and 2) in contrast to a conventional role in gene repression via h2K27me3, Ez, in its Ser21 phosphorylated form, functions as a coactivator of Tcf1 to induce Bcl6 during Tfh responses to viral infections (Li et al. Nat. Commun. 2018). The Xue lab is investigating molecular determinants that instruct Tfh fate decision in CD4+ T cells activated by pathogen infection and protein immunization.
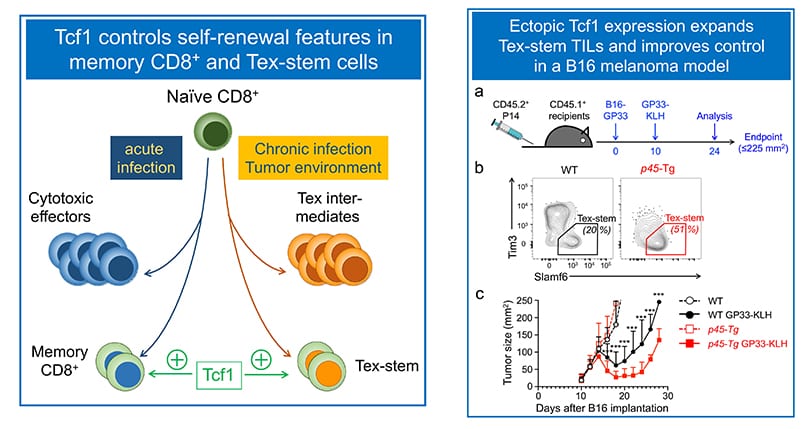
Mapping pathways that program self-renewal in antigen-experienced cytotoxic T cells:
Mature CD8+ T cells are cytotoxic toward cells infected by viruses and intracellular pathogens, as well as malignantly transformed cells. Recent studies revealed CD8+ T cell plasticity tailored to the nature of infectious agents or tumor microenvironment. In response to acute viral or bacterial infections, activated CD8+ T cells give rise to cytotoxic effector cells that clear the pathogens, and memory CD8+ T cells that confer long-term protection against the pathogens. We have established a critical requirement for Tcf1 for persistence of memory CD8+ T cells and their recall responses (Zhou et al. Immunity, 2010. In the context of chronic viral infection and tumor microenvironment, however, CD8+ T cells become dysfunctional/exhausted, known as Tex cells. In collaborative work with the Ahmed lab (Emory), we contributed to the discovery that a subset of Tex cells possess stem-cell like features (called Tex-stem cells), showing longer persistence and enhanced proliferative capacity in response to anti-PD1 checkpoint blockade (Nature 2016). Our recent studies have demonstrated that Runx3 antagonizes in Tex-stem cell formation (Shan et al, Nat Immunol. 2017, and ectopic Tcf1 expression instills the Tex-stem program to enhance viral and tumor immunity (Shan et al, Cell Mol Immunol. 2020. The Xue lab aims to identify key pathways that enhance recall responses by memory CD8+ T cells or rectify T cell exhaustion using infection and tumor models.