Research in the Ryu Lab
Cancer Cell Epigenome Reprogramming by Targeting Chromatin Remodeling Proteins
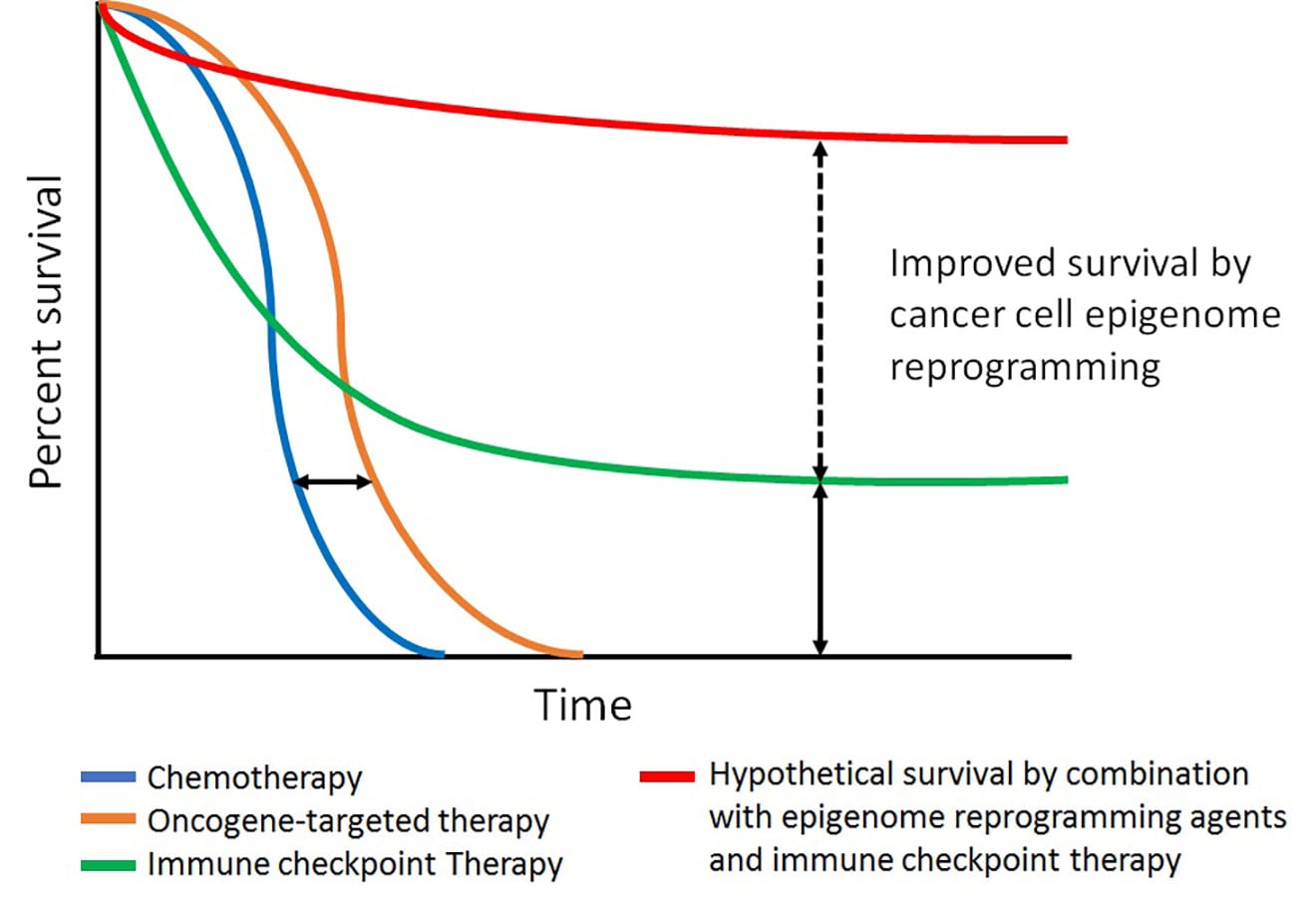
Kaplan-Meier survival curves of currently available therapies for melanoma. Improved overall survival (OS) but lacking durable benefit of oncogene-targeted therapy (orange line) compared with standard chemotherapies (blue line). Immune checkpoint therapies (green line) show long-term clinical benefit compared with oncogene-targeted therapies, but only in a fraction of patients. Combination with epigenetic agents and immune checkpoint therapies (red line) may improve OS rate compared to immune checkpoint therapy alone.
Recent advances in basic and clinical research have led to the development of therapies for melanoma patients, providing significant benefit. One is oncogene-targeted therapy, which shows a high rate of initial clinical response in patients with the tumors containing a mutated oncogene, but clinical benefits are short-lived due to the rapid onset of resistance to therapy in most patients. The second line of therapy is immune checkpoint blockade, which empowers antitumor T-cell responses in patients; this approach results in a durable long-term clinical benefit, but only in a limited number of patients. In this project, our research focuses on increasing cancer cell susceptibility to these therapies by epigenetically reprogramming melanoma cells through the manipulation of chromatin remodeling enzymes—such as ATPase-dependent SWI/SNF proteins, class I histone deacetylases (HDAC I and II) containing transcriptional co-repressor complexes, and protein complexes interacting with DNA methyl-transferases—in the context of existing therapies to increase clinical benefit. To achieve this goal, we use unbiased tools, such as genome/epigenome-wide association study, chemical genetics approaches, and animal models.
Kalin JH, Wu M, Gomez AV, Song Y, Das J, Hayward D, Adejola N, Wu M, Panova I, Chung HJ, Kim E, Roberts HJ, Roberts JM, Prusevich P, Jeliazkov JR, Roy Burman SS, Fairall L, Milano C, Eroglu A, Proby CM, Dinkova-Kostova AT, Hancock WW, Gray JJ, Bradner JE, Valente S, Mai A, Anders NM, Rudek MA, Hu Y, Ryu B, Schwabe JWR, Mattevi A, Alani RM, Cole PA. Targeting the CoREST Complex with Dual Histone Deacetylase and Demethylase Inhibitors. Nature Commun. 2018 Jan 4;9(1):53. PMCID: PMC5754352
Yan G, Eller MS, Elm C, Larocca CA, Ryu B, Panova IP, Dancy BM, Bowers EM, Meyers D, Lareau L, Cole PA, Taverna SD, Alani RM. Selective Inhibition of p300 HAT Blocks Cell Cycle Progression, Induces Cellular Senescence, and Inhibits the DNA Damage Response in Melanoma Cells. J Invest Dermatol. 2013 Oct; 133(10):2444-52. PMCID: PMC4380234.
Ryu B, Fletcher TM, Baumann CT, Warren BS, Fragoso G, John S, Hager GL. Structure and dynamic properties of a glucocorticoid receptor-induced chromatin transition. Mol Cell Biol. 2000 Sep; 20(17):6466-75. PMCID: PMC86121.
Novel Strategies for Functional Reactivation of Tumor Suppressors in Melanoma
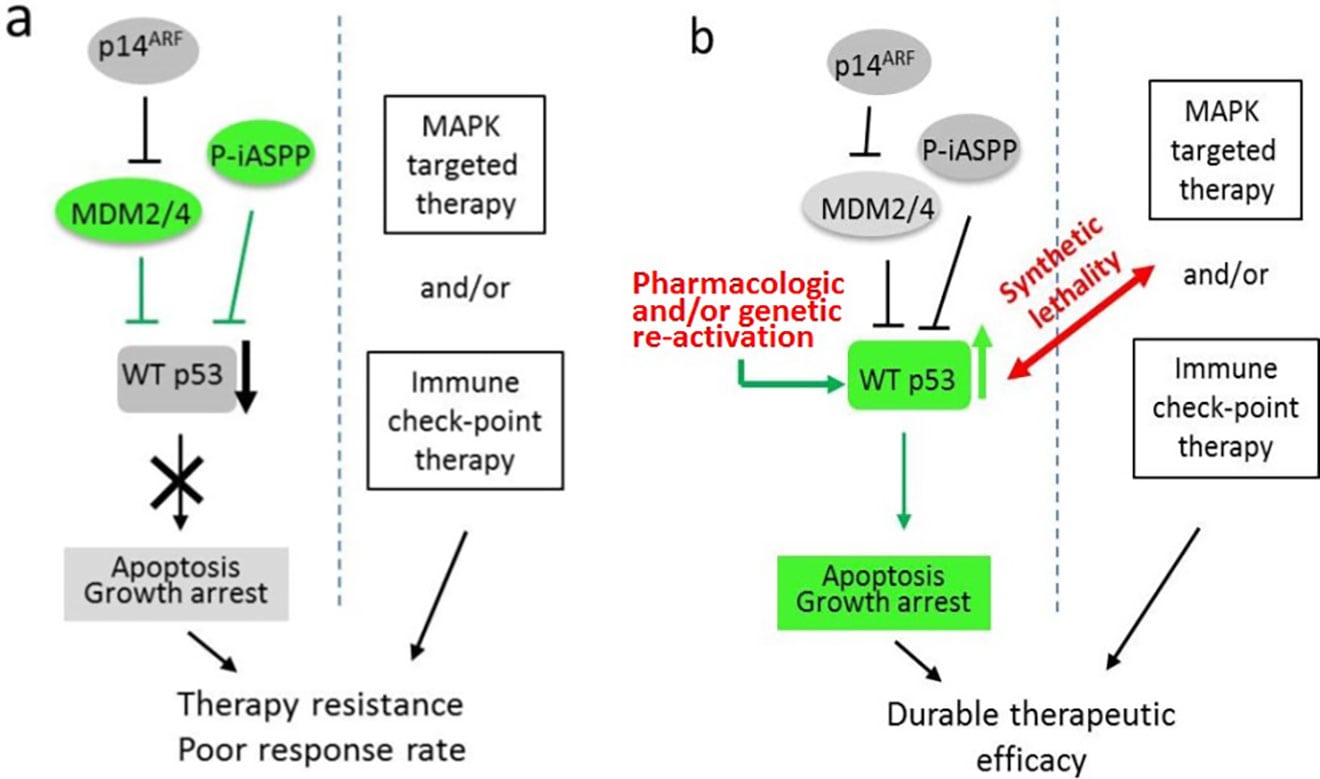
Schematic diagrams of synthetic lethality by p53 reactivation in melanoma therapy. (a) Targeted therapy with WT p53 inactivation resulted in poor therapeutic effect. (b) Induction of synthetic lethality by WT p53 reactivation in conjunction with FDA-approved targeted therapies may result in increased therapeutic efficacy.
Mutation frequencies of several tumor suppressor genes in melanoma, including Tp53 and PTEN, are dramatically lower compared to other solid tumors. Hence, patients with wild-type tumor suppressor genes could benefit from reactivation of tumor suppressive pathways. However, many studies have demonstrated that tumor suppressors are functionally repressed by various regulatory mechanisms. It is highly likely that there are other uncharacterized molecular mechanisms of functional inactivation in melanoma, such as epigenetic silencing and/or post-translational modifications (PTMs). Hence, uncovering those mechanisms could lead to new approaches to reactivate tumor suppressors as a treatment for melanoma. Our future research is focused on developing novel strategies to reactivate those proteins using chemical inhibitors of lysine-modifying enzymes in the context of synthetic lethality of oncogene-targeted and immune checkpoint therapies.
Su GH, Sohn TA, Ryu B, Kern SE. A novel histone deacetylase inhibitor identified by high-throughput transcriptional screening of a compound library. Cancer Res 2000 Jun 15; 60(12): 3137-42. PMID: 10866300.
Molecular Signatures and Biomarker Discovery for Melanoma
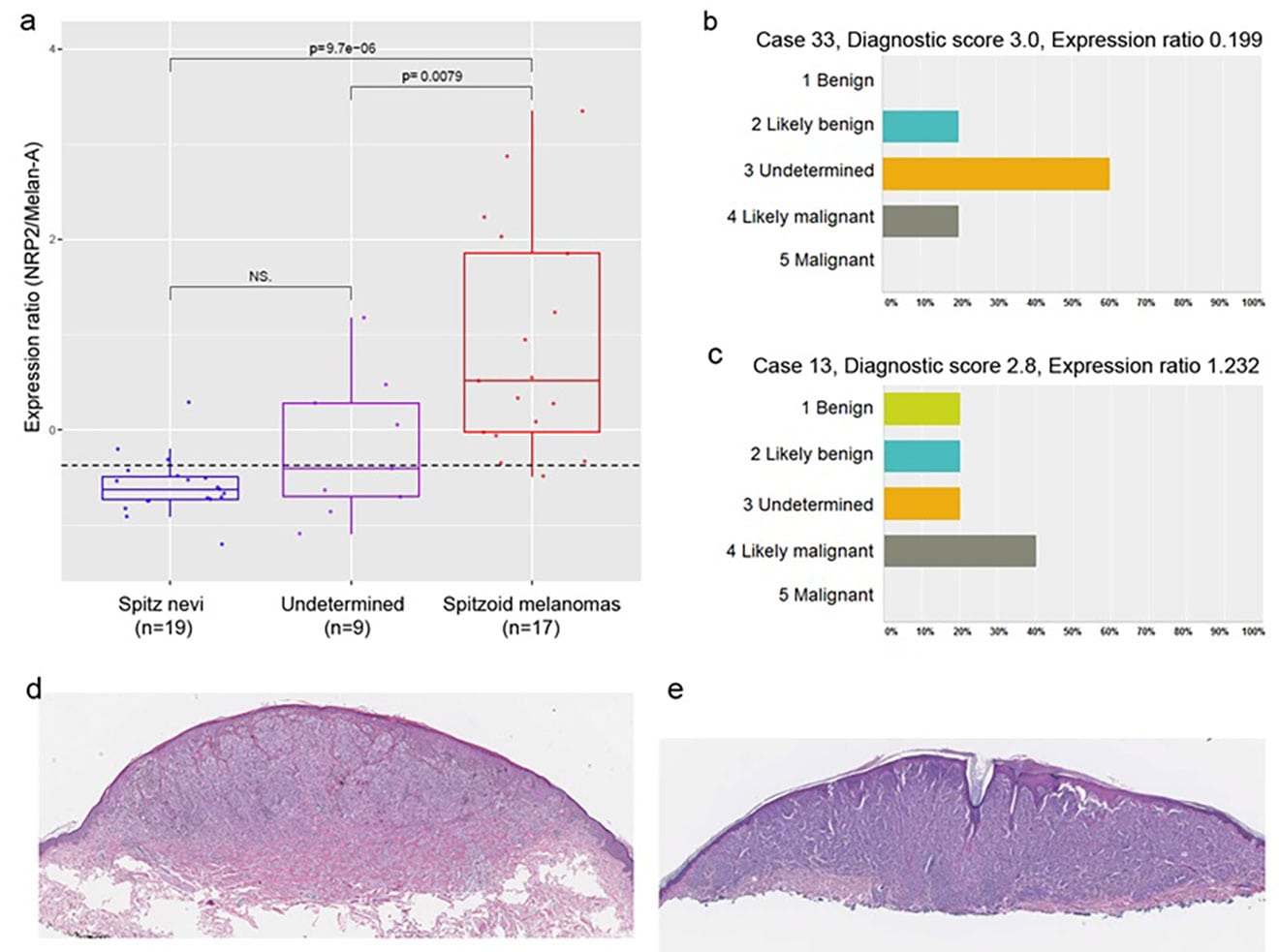
Identification of spitzoid melanocytic lesions and their correlations with values of NRP2/melan-A gene expression ratios. (a) Histologic re-examination of tissue specimens by five independent dermatopathologists identifies three groups of samples: Spitz nevi (SN), spitzoid melanoma (SMs), and atypical spitzoid neoplasms (undetermined). Box–Whisker plot analysis of the NRP2/melan-A expression ratio of the three sample groups shows complete segregation between SN and SMs. (b, c) Two representative cases (cases 13 and 33). Case 33 could be classified as likely benign Spitz nevus based on the diagnostic NRP2/melan-A expression ratio, whereas case 13 could be categorized as likely malignant spitzoid melanoma. (d, e) Hematoxylin and Eosin stained section of Spitz nevus (case 33) and spitzoid melanoma (case 13), respectively. Eisenstein A and Ryu B et al., Melanoma Res. 2018 Feb;28(1):71-7.
Current practice in the prognostic evaluation of melanoma largely relies on macroscopic histopathological features of affected lesions and immunohistochemical (IHC) analysis of tissue samples. These prognostic methods should be improved in terms of accuracy and ease of use in clinical settings for patient classification and personalized treatment. We are using genome-wide association studies for expression profile and DNA methylation analysis to identify molecular signatures associated with malignant cancer progression and to discover predictive biomarkers of prognostic and therapeutic outcome for melanoma.
Eisenstein A, Panova I, Chung HJ, Goldberg LJ, Zhang Q, Lazova R, Bhawan J, Busam KJ, Symanowski JT, Alani RM, Ryu B. Quantitative assessment of Neuropilin-2 as a Simple and Sensitive Diagnostic Assay for Spitzoid Melanocytic Lesions. Melanoma Res. 2018 Feb;28(1):71-7
Rossi M, Tuck J, Kim OJ, Panova I, Symanowski JT, Mahalingam M, Riker AI, Alani RM, Ryu B. Neuropilin-2 Gene Expression Correlates with Malignant Progression in Cutaneous Melanoma. Br J Dermatol. 2014 Aug;171(2):403-8. PMID: 24359286.
Kim HE, Symanowski JT, Samlowski EE, Gonzales J, Ryu B. Quantitative measurement of circulating lymphoid-specific helicase (HELLS) gene transcript: a potential serum biomarker for melanoma metastasis. Pigment Cell Melanoma Res. 2010 Dec; 23(6):845-8. PMCID: PMC3192016.
Ryu B, Kim DS, Deluca AM, Alani RM. Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PLoS One. 2007; 2(7):e594. PMCID: PMC1895889.
